Oral Presentation 16th Lorne Infection and Immunity 2026
Investigating the genotype dependence of viral infection and immune phenotypes using genetically diverse human cell libraries (#6)
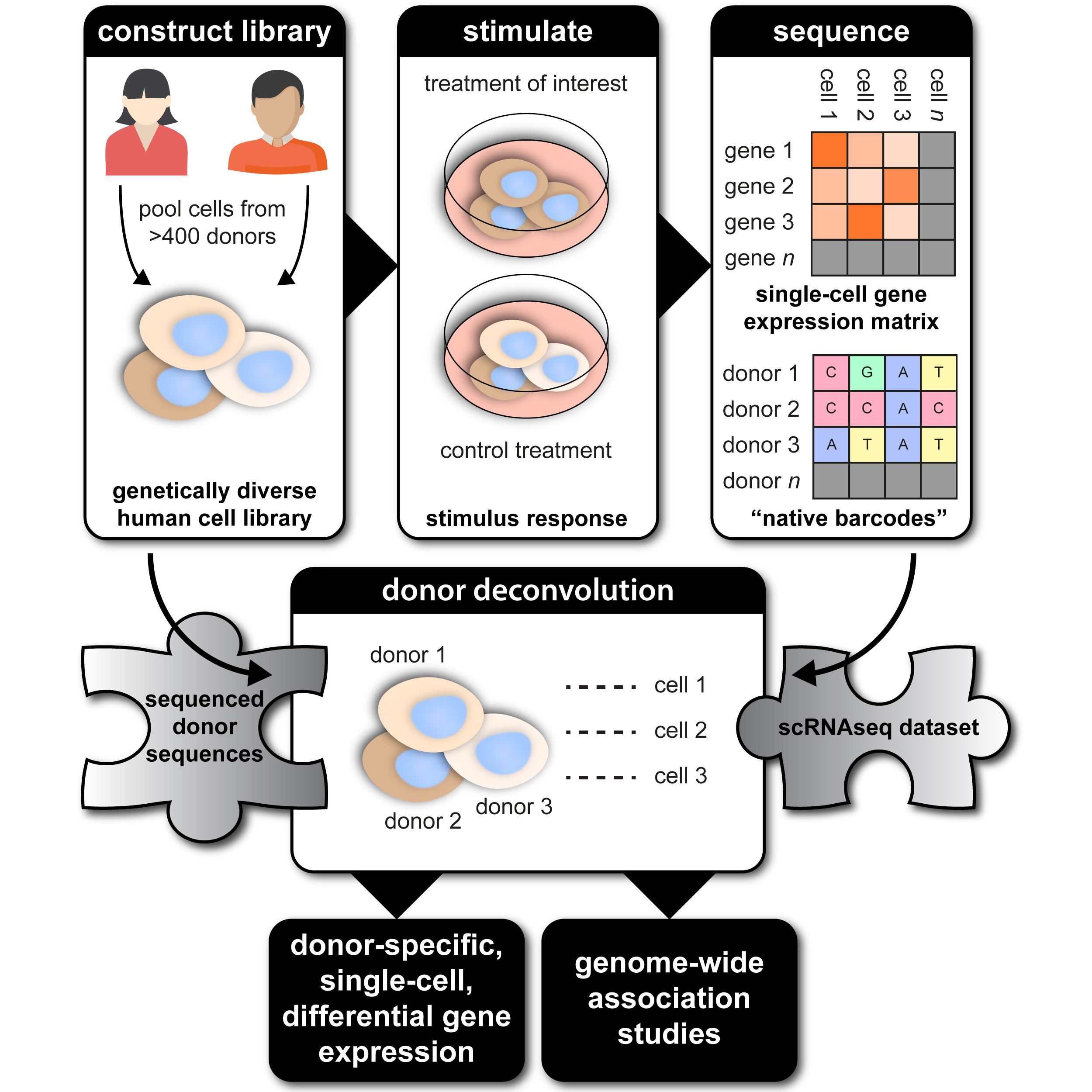
Genetic variation between populations, individuals, and even individual cells drives differential responses to stimuli, complicating the analysis and prediction of stimulus phenotypes across biological scales. Classically, this variation is controlled using cultured cells or other model organisms drawn from a limited set of genetic backgrounds, which is typically unrepresentative of the diversity of phenotypic responses in actual populations. Through somatic recombination of immunoglobulin genes, the vertebrate adaptive immune system harbors a still greater source of genetic diversity. Here, we leverage the recombined immunoglobulin gene repertoire as a matrix of “native genetic barcodes” to assign cells within heavily mixed pools to their donor-of-origin, and then each donor to their personal stimulus phenotype after treatment. Using our novel approach, we uncover genotype-dependent responses to Toll-like receptor (TLR) agonists, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) gene expression, and chronic Epstein–Barr virus (EBV/HHV-4) infection. Our data challenges the hypothesis that the SARS-CoV-2 ORF3a protein generates cytotoxic viroporins, but may instead act as a virulence factor that stimulates hyperinflammation associated with severe coronavirus disease (COVID) in a defined subset of individuals. Our experimental method and deconvolution algorithm leverage genetic diversity to democratize and accelerate the analysis of genotype–phenotype relationships for virtually any stimulus of interest.
- Briney et al. (2019), "Commonality despite exceptional diversity in the baseline human antibody repertoire", Nature, 566(7744):393-7.
- Kern et al. (2021), "Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs", Nature Structural and Molecular Biology, 28:573-82.
- Sirugo et al. (2019), "The missing diversity in human genetic studies", Cell, 177:26-31.
- Ren et al. (2020), "The ORF3a protein of SARS-CoV-2 induces apoptosis in cells", Cellular and Molecular Immunology 17:881-3.